-
Notifications
You must be signed in to change notification settings - Fork 60
Description
We've known about this for a while now, I think it's time to at least make it an official feature-request. A solution is not urgent, though.
Matrices which are used in ODEPrior are, more often than not, of the form np.kron(mat1, mat2) usually even np.kron(np.eye(dim), mat). This allocation eats too much storage and has a slow MVM.
I propose to use a data structure such as for instance BlockDiagonal which only stores mat and dim and implements fast matrix-vector products:
class BlockDiagonal(pn.linalg.linops.LinearOperator):
"""
Cheap implementation of np.kron(np.eye(num_copies), block).
Optimised for a fast '@' operation.
"""
__slots__ = ['block', 'num_copies']
def __init__(self, block, num_copies):
self.block = block
self.num_copies = num_copies
def toarray(self):
return np.kron(np.eye(self.num_copies), self.block)
def __add__(self, other):
assert self.num_copies == other.num_copies
return BlockDiagonal(self.block + other.block, self.num_copies)
def __sub__(self, other):
assert self.num_copies == other.num_copies
return BlockDiagonal(self.block - other.block, self.num_copies)
def __matmul__(self, other):
if isinstance(other, BlockDiagonal): # matmul
assert self.num_copies == other.num_copies
return BlockDiagonal(self.block @ other.block, self.num_copies)
if isinstance(other, np.ndarray): # matvec
return (self.block @ other.reshape((self.block.shape[1], -1))).T.flatten()
raise NotImplementedError
@property
def T(self):
return BlockDiagonal(self.block.T, self.num_copies)For projection matrices odeprior.proj2coord(), one could even go one step further and implement slicing into matmul (which I have not drafted yet). It would make the code equally readable (in my opinion; it would still use proj @ cov @ proj.T), the step to a more general kronecker product np.kron(mat1, mat2) would remain a simple extension (using e.g. (A \otimes B)(C \otimes D)=(A \otimes C)(B \otimes D); almost all involved matrices have this kronecker structure)) and the memory footprint as well as the speed would improve significantly (see below)
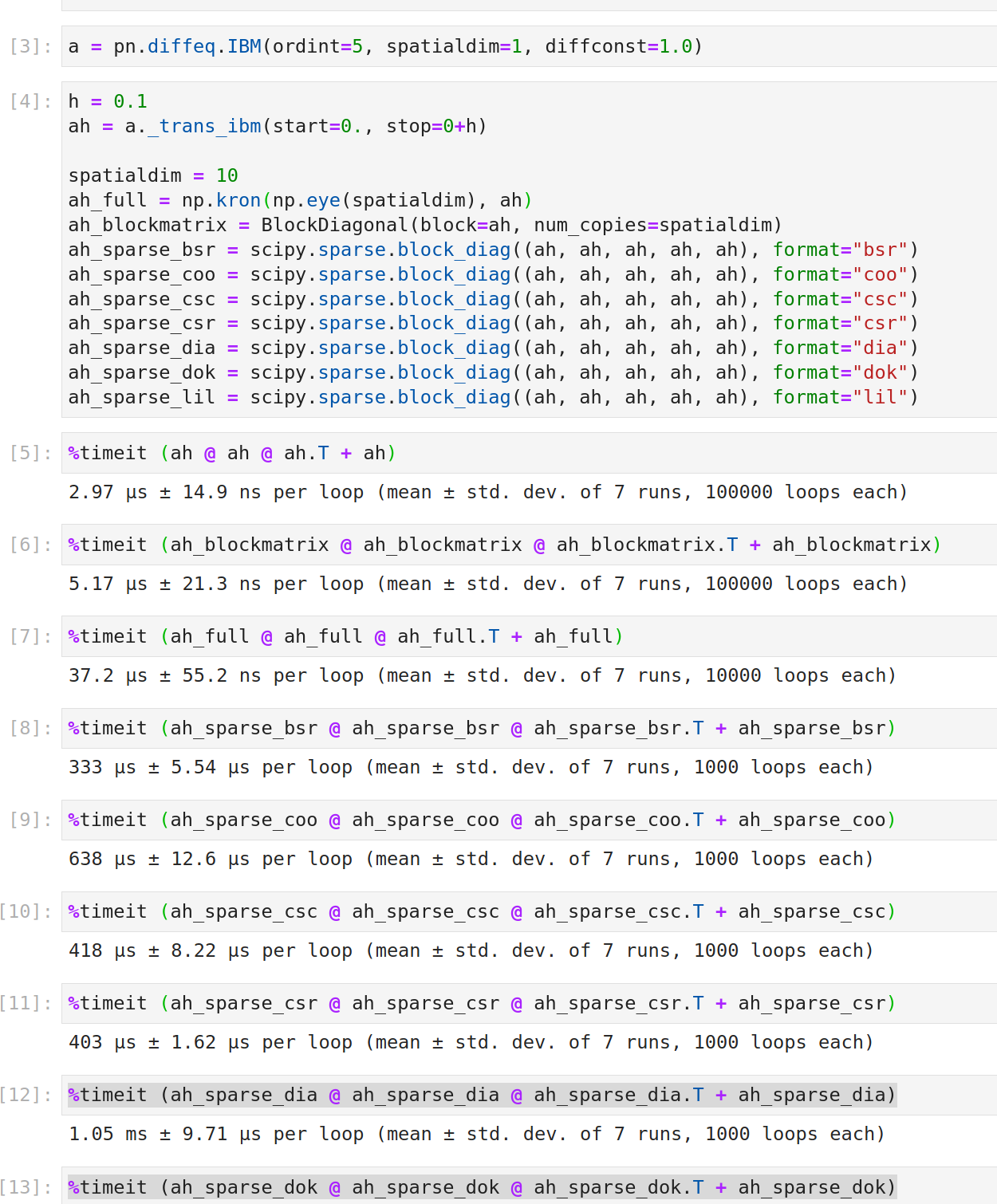
Affected files would be prior.py and ivp2filter.py which would need to replace the respective outputs by the data structure. Occasionally, np.linalg.solve() and np.linalg.cholesky() calls in ivpfiltsmooth.py and gaussfiltsmooth.py would have to be replaced. It could be a smart choice to subclass from linops.LinearOperator for interface reasons.